Perhaps a mobile piece of DNA that moves from one place to another in the bacterial genome could be turned into a precise new genetic engineering tool to develop medical treatments and crops. After decades of research on this mobile piece of DNA, a group of scientists have now explained how it attaches to very specific places in the genome and then integrates there. They were able to insert genes into bacteria via this piece of DNA coli bacteria Or turn off his genes. They describe this in two articles published Wednesday in nature.
“This is a wonderful discovery,” says Marcel Tijsterman, professor of genome stability at Leiden University Medical Center, who was not involved in the study. He praises the “elegance of the work.”
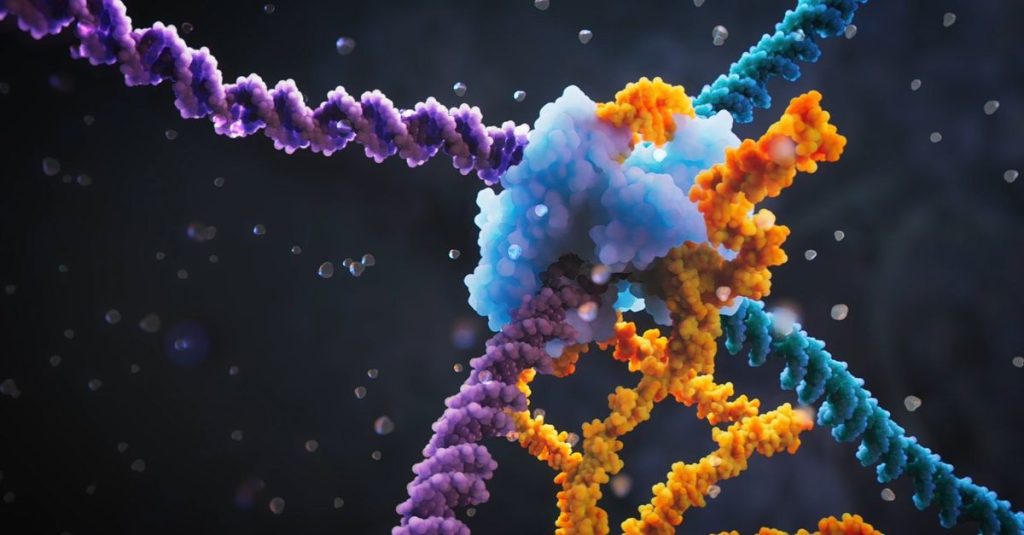
The mobile piece of DNA falls into the category of so-called Movable items, moveable items. It moves itself through an organism’s genome, or makes copies of itself, and that copy then integrates into a different location in the genome. If this happens in a place where the gene is present, that gene can be turned off. These transposable elements can also move or reverse longer stretches of DNA (including genes). They make up up to 40 percent of the total genome of some bacteria, and in maize up to 85 percent. Therefore, they also have a significant impact on the three-dimensional structure of DNA, and therefore also on gene activity.
Biotechnology is eagerly exploiting this mechanism. In recent years, techniques have been developed to insert very precise pieces of DNA into the genome of bacteria, plants or animals. The most famous of these is the CRISPR-Cas technique, which won the 2020 Nobel Prize in Chemistry. The CRISPR component codes for a 20-letter sequence of DNA and binds to a complementary 20-letter code in the genome, after which the cas component cuts the DNA in half at that location. A gene can be placed in between.
Does this technique also work on mammals and plants? This is still the question
The now-deployed mobile DNA fragment is linked to the CRISPR-Cas system, says biochemist Raymond Stahl (Wageningen University and Research), who works on this system. The mobile piece of DNA belongs to the family of the simplest transposable elements: Insertion sequences. These are found in bacteria. The researchers focused on one specific member of that family, element IS110. It cuts itself out of the genome in one place and then integrates itself in another. When IS110 cuts itself, part of it is translated into mRNA and then into a recombination protein. The discovery now is that another part of IS110 is also translated, but remains “stuck” in the RNA phase. Scientists call this piece “RNA bridge“. The RNA bridge binds to the DNA code present in the genome at a specific place, and at the same time binds to IS110. Researchers have been able to visualize these connections – one of the two publications is on this topic. So the RNA bridge ) moves IS110 to a site in the bacterial genome where it can integrate. Recombination ensures this occurs.
The IS110 system is better in some ways than the Crispr-Cas system, Stals says. “I always have to explain that the Crispr-Cas machine works very precisely, but it just makes a cut somewhere in the genome. If you then want to insert genes, you need additional techniques. But the IS110 system does both. Well, It’s doubtful “Whether it also works in mammalian and plant cells.”
Tijsterman has another comment. The code through which the RNA bridge carries out communication is only 8 to 10 letters long. bee coli bacteria There is a small chance of this code being duplicated. But in the much larger genomes of plants or humans, such a short codon occurs more often. “The chance of introducing genes into an unwanted place is greater.” Tegsterman expects the search will now begin for approximately 30 ISIS families. “Maybe there is a family member whose code has more letters.”

“Coffee buff. Twitter fanatic. Tv practitioner. Social media advocate. Pop culture ninja.”










More Stories
Which can cause an increase in nitrogen.
The Central State Real Estate Agency has no additional space to accommodate Ukrainians.
The oystercatcher, the “unlucky national bird,” is increasingly breeding on rooftops.